The earlier two articles have been solely introductions into the primary matter of this knowledgeable collection. On this and the next two articles, we’ll get into the precise matter of epigenetic deregulation throughout getting old. Nonetheless one couldn’t perceive epigenetic deregulation with out first understanding epigenetics. So hopefully by now you’ve gotten a fundamental understanding of what epigenetics is, why it exists and the way it’s maintained and established. On this article we’ll get into what precisely occurs to the epigenome throughout getting old, why it occurs and the way we are able to measure it for our profit. Firstly let’s discuss how epigenetics performs into the plethora of modifications that happen throughout getting old.
This text is an element three of a four-part collection on reproductive getting old.
- Introduction to Epigenetics
- Epigenetics and Gene Expression
- Epigenetics and Growing old
- Coming Quickly
Now should you’ve ever tried to enterprise into the sphere of getting old science the primary article that you just’ll probably bump into or discover generally referenced by nearly all getting old papers is a overview article referred to as the “Hallmarks of getting old”. This text self-admittedly makes an attempt to duplicate the landmark paper “Hallmarks of most cancers” for the sphere of getting old. On this article the authors set up 9 totally different getting old hallmarks that they argue “…are typically thought of to contribute to the getting old course of and collectively decide the getting old phenotype”.1 Amongst these 9 hallmarks is epigenetic alterations. Nonetheless, barely totally different from the opposite hallmarks, epigenetic alterations are among the many hallmarks denoted as “major”. Which means they’re the causes of harm, or locations that harm manifests in. Different classes are, antagonistic (responses to wreck) and integrative (outcomes of cumulative harm). The first hallmarks are arguably extra necessary within the sense that their deregulation precedes the opposite hallmarks and as a rule causes them. Interventions that focus on the first hallmarks may additionally alleviate the signs that come up from the opposite hallmarks.
Determine 1: The 9 key hallmarks of getting old as mentioned within the overview: genomic instability, telomere attrition, epigenetic alterations, lack of proteostasis, deregulated nutrient sensing, mitochondrial dysfunction, mobile senescence, stem cell exhaustion, and altered intercellular communication. Sourced from Lopez-Otin et al.1
Epigenetics arguably holds an much more necessary place among the many different major hallmarks of getting old due to its analog, complicated nature. The opposite major hallmarks embody genomic harm, telomere attrition and lack of proteostasis. Whereas genomic harm and telomere attrition affect the mobile equipment in a comparatively direct method, epigenetic alterations function by means of a extra nuanced and multifaceted pathway, influencing gene expression and mobile id with out altering the underlying DNA sequence. This analog and sophisticated mechanism permits epigenetics to exert a broad affect over the organic processes that dictate getting old, appearing as a grasp regulator of the mobile response to getting old and environmental influences.
Modifications in epigenetic modifications throughout getting old
The intricate system that’s the epigenome, which incorporates DNA methylation, histone modifications, non-coding RNAs, and chromatin reworking, is basically the maestro of gene expression. With age, this conductor begins altering its tune, resulting in shifts in how genes carry out. This exploration goals to unpack the nuances of those epigenetic modifications and their vital function within the getting old course of. Earlier than going onto the precise modifications and the way they modify throughout getting old it might be higher to speak in regards to the epigenome as a complete. One frequent phenomena noticed constantly throughout getting old is the erosion of heterochromatin throughout getting old. As talked about within the earlier article heterochromatin is the tightly compacted, typically not expressed elements of chromatin. They naturally differ from cell kind to cell kind. Preserving genes and pathways that aren’t required silenced. With age the epigenetic marks that denote heterochromatin places start to weaken, resulting in a much less compact and extra transcriptionally energetic state. This lack of heterochromatin integrity permits genes which are usually silenced to grow to be erroneously expressed, contributing to the mobile dysfunction usually seen in getting old. This understanding of heterochromatin erosion units the stage for analyzing particular epigenetic modifications corresponding to DNA methylation, histone modifications, and non-coding RNA interactions, and their roles within the getting old course of.
Transcriptional modifications
Transcriptional output, the eventual results of the epigenetic panorama is how age-related epigenetic modifications manifest themselves within the phenotype. Quite a few research have recognized modifications within the expression ranges of particular person genes and full pathways related to getting old. For instance, a research of gene expression profiles of practically 15000 people identifies round 1500 genes that change considerably with age.2 The genes present lowered exercise in RNA and protein synthesis, mitochondrial features, and DNA restore, alongside an upregulation in immune responses and metabolic processes throughout the research.
Nonetheless, it’s necessary to grasp that getting old impacts every individual otherwise, making it a really various course of. Think about making an attempt to grasp what a typical household appears to be like like primarily based on just some examples; with solely three youthful and three older people’ information, it’s powerful to seize the complete vary of variations that may exist. Simply as households differ extensively in how many individuals they’ve, what they love to do, or the place they stay, the modifications in gene exercise as we age can even differ significantly from one individual to a different.
This selection implies that when researchers solely have a look at a small variety of individuals, they’ll’t confidently say what typical aging-related modifications would possibly appear like. They want lots of information from many various individuals to see the actual patterns of how getting old impacts gene exercise. Due to this fact, whereas there are basic tendencies in how getting old impacts our genes, pinpointing these tendencies in small research may be unreliable as a result of excessive stage of pure variation in gene expression amongst people.
DNA methylation
The connection between DNA methylation and getting old, though one of many easiest amongst epigenetic modifications, continues to be considerably complicated. DNA methylation, a key epigenetic change, entails the addition of methyl teams to particular DNA areas, primarily at cytosine bases in CpG dinucleotides. This course of is essential for regulating gene exercise and performs a major function within the getting old course of. As we age, a basic development of DNA methylation loss is noticed.34 This can be attributable to the modifications within the exercise ranges of the enzymes chargeable for DNA methylation, often called DNA methyltransferases (DNMTs). DNMT1, the variant chargeable for upkeep of present DNA methylation marks tends to lower, decreasing general methylation ranges and doubtlessly resulting in elevated gene exercise that was beforehand suppressed.56
Whereas world DNA methylation ranges are inclined to lower as we age, we are able to additionally observe particular and distinct modifications inside the methylome—the entire set of DNA methylation in our cells. Some websites exhibit achieve of methylation with age whereas some websites lose DNA methylation distinct from the worldwide loss.78 Furthermore, these methylation modifications aren’t simply random; some are constant throughout totally different species, suggesting a standard organic getting old phenotype.9 Actually we try to uncover and quantify these non-random modifications in DNA methylation with machine studying by means of the usage of epigenetic clocks, which we’ll get into later within the article.
Histone associated modifications
Histone modifications and modifications in histone variants are essential to understanding the epigenetic mechanisms that affect how we age. These processes contain tweaks to the proteins round which DNA is wrapped, affecting gene exercise. As we get older, particular modifications in these histone proteins and their modifications can affect how cells operate and contribute to the getting old course of.
Histone variants are particular variations of the usual histone proteins that play distinctive roles in DNA and gene regulation. Not like common histones, that are changed throughout DNA replication, variants like H3.3 are added to chromosomes independently of replication. That is necessary for sustaining chromatin construction in cells that not divide, corresponding to these in getting old tissues. As an example, H3.3 turns into the primary type of histone H3 in senescent human cells, highlighting its function in mobile getting old.1011 Moreover, the variant macroH2A, which will increase with age, is linked to repressing genes related to mobile getting old and performs a essential function in preserving the chromatin organized throughout senescence.12
Histone modifications—like including or eradicating methyl or acetyl teams—can both loosen or tighten DNA packaging, thus influencing whether or not genes are turned on or off. For instance, acetylation normally opens up the chromatin to advertise gene exercise, whereas methylation can both activate or silence genes relying on the place and what number of methyl teams are added. Acetylation at particular websites on histones H3 and H4 (H3 K56Ac and H4 K16Ac) can affect getting old in several methods. In yeast H3 K56Ac is linked to improved DNA stability and restore, selling longevity.13 Alternatively, a rise in H4 K16Ac in older cells is related to getting old processes, indicating {that a} correct stability of acetylation is essential for an extended life.14 Methylation modifications, particularly the trimethylation marks on histones, additionally play a major function in getting old. Extra methylation at sure websites would possibly silence genes that have to be energetic for youthfulness, contributing to age-related decline, whereas much less methylation would possibly activate genes that result in getting old.15 Modifications in histone methylation with age mirrors the lack of heterochromatin, that means we observe a rise in energetic histone methylation marks whereas a lower in repressive histone marks.1617 Actually this development was even proven to be causal within the nematode worm C. elegans. Decreasing the energetic chromatin mark H3K4me3 by lowering the expression of the proteins that deposit that modification led to a rise in lifespan and conversely the lower of proteins that take away this mark decreased lifespan.18
Modifications in histone marks, which are available many types, point out a lack of heterochromatin. But, these modifications don’t neatly present the distinction between young and old cells. Whereas we’ve pinpointed particular modifications that may affect lifespan, there’s nonetheless a lot we don’t totally perceive. Moreover, these findings have to be confirmed in mammals and people to make sure they’re relevant extra broadly. Though our data has gaps, it’s clear that histone modifications play an important function in getting old and illness.
nc-RNAs
We’ve got explored nc-RNAs and their features beforehand. As soon as part of the genetic materials labeled as “junk DNA”, the physique of proof supporting their roles as key regulators in genetic networks is ever rising. The analysis on the altering dynamics of nc-RNAs throughout getting old are far much less understood in comparison with different epigenetic modifications. Nonetheless there’s a lately rising physique of literature that elucidates their modifications in getting old.
The enzyme Dicer, which performs a key function in reducing small non-coding RNAs, corresponding to miRNAs, into their energetic types, reveals much less exercise in older organisms.19 This means a basic lower in ncRNA regulation with age. MiRNAs themselves, which fine-tune gene expression by focusing on mRNAs, endure modifications in expression as organisms age. They dramatically have an effect on lifespan and tissue getting old, as demonstrated in research with C. elegans the place sure miRNAs improve longevity whereas others shorten it.20 Like most early analysis areas most miRNA – getting old relationships have been proven in decrease organisms. Among the many few which were proven to have an effect on lifespan in mammalian fashions there’s miR-17. Mice expressing miR-17 noticed a 16% improve in lifespan together with decreased mobile senescence.21
Alternatively, lengthy non-coding RNAs (lncRNAs) additionally considerably affect getting old by interacting with chromatin and protein complexes, affecting every thing from genomic stability to irritation. For instance, increased ranges of the lncRNA Gas5 in aged mouse brains have been linked to impaired studying talents.22 Equally, H19, one other lncRNA, performs a task in regulating gene networks that might result in age-related illnesses like prostate most cancers when altered.23
But, regardless of these insights, our understanding of how ncRNAs regulate mobile processes, together with getting old, stays incomplete. This hole underscores the necessity for additional analysis to completely grasp and doubtlessly harness ncRNAs in getting old.
Chromatin Remodelling and 3D genome construction
Chromatin, the complicated of DNA and proteins present in our cells, isn’t randomly bunched into the nuclei of cells however reasonably is in an organized three-dimensional construction that’s essential for regulating gene exercise. This three-dimensional construction separates the genetic materials into compartments for organized epigenetic management and might differ from cell to cell. Nonetheless, like all epigenetic mechanisms it is usually topic to deregulation with age. Sequencing applied sciences that enable to seize 3D interactions within the genome are comparatively latest which makes this space a newly growing discipline. A lot of the data current overlaps with the acquisition of senescence reasonably than explicitly getting old. For instance, in senescent human mesenchymal stem cells and fibroblasts, areas marked by H3K27me3 swap from inactive (B) to energetic (A) compartments, altering gene expression linked to getting old.24

Determine 2: As getting old progresses, there’s a basic lack of heterochromatin and a detachment of lamina-associated domains (LADs) from the nuclear lamina. This restructuring of higher-order chromatin is accompanied by shifts in histone modifications, resulting in elevated chromatin accessibility, activation of repetitive sequences, and dysregulated gene expression. Sourced from Wang et al.25
Epigenetic data upkeep
So epigenetic data is misplaced with getting old. We went over a wide range of modifications that change in direction of a selected path throughout getting old. There should exist drivers of this transformation. Phenomena that result in lack of epigenetic data and there have to be a system in cells to oppose this transformation. A system that goals to take care of and propagate epigenetic data in opposition to these “deregulators”. Wherever/each time the upkeep system comes quick some epigenetic data is misplaced and contributes to the buildup of loss over the lifespan of an organism which in flip contributes to getting old.
Epigenetic data within the cell cycle
Epigenetic inheritance throughout cell division is essential for sustaining mobile id and performance throughout generations. This course of ensures that epigenetic data, corresponding to DNA methylation and histone modifications, is precisely transmitted from mom cells to daughter cells, regardless of the disruptive nature of cell replication.26 Throughout cell division, precisely replicating DNA’s genetic and epigenetic data is essential. DNA methyltransferase enzymes play a key function by copying methylation patterns from the father or mother DNA to the brand new strands, making certain that DNA methylation—which usually suppresses gene expression—is preserved in daughter cells.27 Equally, histone modifications corresponding to methylation and acetylation, which affect chromatin construction and gene exercise, are meticulously replicated. Previous histones are redistributed between the brand new DNA molecules, and new histones are added and modified to mirror the unique patterns, guided by the unique histone ‘template’.28
Filling within the blanks
The epigenome, a fancy community of modifications that cells make the most of throughout each growth and regular functioning of terminally differentiated cells, displays a major characteristic: the crosstalk between totally different modifications. These relationships are intricate and multifaceted. As an example, it’s well-documented that DNA methylation usually co-locates with a selected histone modification, H3K9me3.29 One other detailed instance reveals that two totally different histone modifications may be related by means of a transcription issue that acknowledges one modification and establishes the opposite.30
Such examples are considerable throughout the epigenome. These interactions are what make Waddington’s epigenetic panorama analogy notably apt. On this mannequin, epigenetic states are akin to a ball’s place on a panorama, settling naturally into valleys which characterize steady states. Because of the reinforcing nature of those interactions, a mere alteration of a single modification is mostly inadequate to shift an epigenetic state. The encompassing epigenetic structure compensates for this transformation, pulling the state again to its unique ‘valley’ by means of the affect of present modifications. To successfully alter an epigenetic state, a number of modifications have to be focused concurrently throughout totally different genomic places. This introduces a stage of redundancy to the epigenome, making certain stability and resilience in gene regulation.
Drivers of epigenetic deregulation
Okay so the epigenome modifications with age. We all know just a little about these modifications. A few of the websites the place they occur and wherein circumstances they occur. However why? Is it deliberate or does it simply occur? We should perceive the mechanisms and pathways that drive epigenetic deregulation to establish interventions that may halt or reverse it.
Exterior / inside harm – basic loss with time
That is probably the most elementary of all the varied drivers of epigenetic data loss. Principally what it quantities to is entropy. The epigenome is an ordered assemble that incorporates data. The medium wherein this data is saved is topic to the pure processes of decay and disruption over time. There are inside and exterior damages that the epigenome amongst different issues within the cell has to outlive. Inner damagers like mobile irritation and metabolic byproducts can disrupt epigenetic markers, altering gene expression and mobile habits.31 Exterior damagers, corresponding to ultraviolet (UV) radiation, environmental pollution, and reactive oxygen species (ROS), additional problem the epigenome’s integrity.3233 These elements introduce errors and modifications to the DNA and related proteins, resulting in a gradual degradation of the epigenetic panorama although single epigenetic mutations termed “epimutations”.34 This entropy not solely impacts the operate of particular person cells however can even contribute to broader organismal getting old and the onset of illness.
DNA replication and cell divisions
One of many most important challenges the epigenome has to beat is the compaction of chromatin throughout cell division. The cell has quite a few established pathways to transmit epigenetic data to daughter cells as talked about above. Regardless of these refined mechanisms, the method isn’t foolproof. Errors can happen, DNA methylation patterns may be incompletely copied throughout cell division, resulting in methylation loss.35 Equally, errors within the placement of histone modifications, regulatory proteins, and RNAs may additionally happen. These errors can accumulate over successive cell divisions, resulting in modifications within the epigenetic panorama that will alter gene expression and mobile operate.36 This gradual epigenetic drift contributes to getting old and varied illnesses, highlighting the complicated and considerably weak nature of mobile inheritance.
DNA harm response
There’s one factor within the cell whose upkeep is unarguably extra necessary for normal mobile operate than epigenetic data and that’s the genetic data. Thus, the cell has a really speedy and acute answer to DNA harm. Upon DNA harm equipment to restore the harm is recruited to the harm web site inside minutes.37 Among the many DNA restore equipment that’s recruited to the harm web site there are chromatin remodelers. As may be understood from the title their features are to rework chromatin for any subsequent processes. Nonetheless within the haste of repairing genetic harm the cell loses epigenetic data on the web site of DNA harm.38
Actually this DNA harm response may end up in a lot epigenetic data loss that when DNA harm is artificially induced within the cells of an organism they exhibit accelerated getting old. Researchers use an enzyme that cuts the mouse genome at 20 totally different websites 19 of which don’t code for proteins. Expressing this protein which results in these DNA cuts ends in a relentless DNA harm response which recruits chromatin remodelers to the harm web site and results in epigenetic data loss. These mice, that are termed “inducible modifications to the epigenome” (ICE) mice, exhibit elevated hallmarks of getting old each at a mobile and physiological stage.39
Transposable components
Transposable components are genetic components that kind a serious a part of eukaryotic genomes. They comprise about half of the mammalian genome. They’re one facet of an historical evolutionary battle to outlive. Transposable components are nucleic acid sequences that may transfer or copy themselves to new positions inside the genome. Due to this capacity they’re generally known as “leaping genes”. They hijack mobile transcription equipment to hold out their wants. These transposable components are considerable and really previous. Thus, by means of evolution our biology has discovered methods to make the most of them for our functions as properly. For instance sure transposable components play an necessary function in growth. Thus, not all are transposable components are equal.40
Nonetheless, just like the DNA harm response equipment, their motion of reducing / copying and pasting themselves into the genome could result in the lack of some epigenetic data by means of the transforming of chromatin. A lot of the transposable components are silenced after growth by means of the attachment of heterochromatin (silenced chromatin) markers.41 Nonetheless with age these markers are eroded resulting in the expression of some transposable components. Their expression results in collateral harm, together with the lack of epigenetic data. This loss is partly attributable to the induction of the DNA harm response, which in flip contributes additional to the erosion of epigenetic data.42 Actually the inhibition of a selected kind of transposable factor led to the reversal of assorted hallmarks of getting old in mouse and human cells with accelerated getting old syndromes. As well as stopping the actions of this transposable factor led to will increase in lifespan of mice with an accelerated getting old syndrome.43
Epigenetic Drift
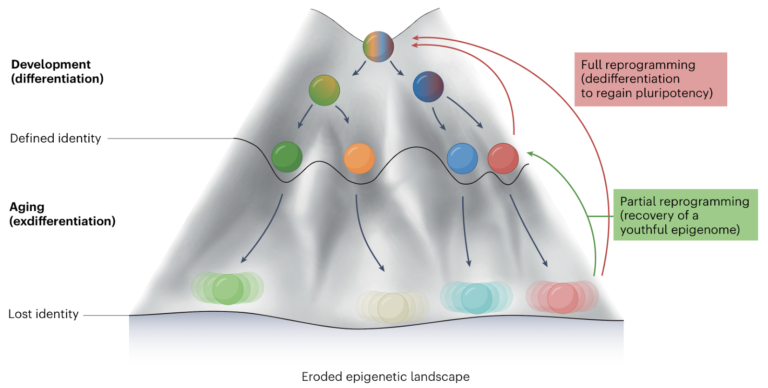
Over time, the mixed results of assorted elements that disrupt epigenetic regulation progressively blur the as soon as clear and particular epigenetic states of cells, main to what’s often called epigenetic drift. This drift outcomes from the buildup of small modifications in epigenetic modifications that happen with out a particular plan, making them unpredictable and difficult to measure. Due to this drift, as cells age, they begin to wander from their ultimate epigenetic states. This epigenetic drift may be prevented by focusing on the drivers of epigenetic deregulation and may be even reversed with interventions corresponding to partial reprogramming as may be seen from determine 3.

Determine 3: The epigenetic panorama, beginning with pluripotent cells on the peak and progressing to differentiated states by means of growth, dictated by modifications like DNA methylation and histone modifications. As getting old happens, DNA harm and different stresses alter this panorama, resulting in cell id drift and practical decline, whereas partial epigenetic reprogramming reveals potential in restoring youthful traits and lengthening lifespan. Sourced from Lu et al.44
As cells deviate from their optimum epigenetic states resulting from epigenetic drift, the consequences of getting old grow to be more and more diverse. This variability is obvious not solely throughout totally different people but additionally among the many cells inside a single individual. Such variety within the getting old course of introduces vital challenges in understanding and monitoring how we age, complicating efforts to establish constant organic markers or develop uniform therapies for age-related circumstances. Given this complexity, there’s a clear want for stylish instruments that may measure and analyze the multifaceted nature of epigenetic modifications. Such instruments would assist us quantify the affect of getting old on the epigenome and assess the effectiveness of assorted interventions geared toward mitigating age-related modifications.
Epigenetic Clocks: An Try at Measuring & Monitoring Epigenetic Deregulation
One try and deconvolute the modifications within the epigenome right into a quantifiable measure are epigenetic clocks. These clocks assist us quantify the complicated modifications taking place inside our epigenome as we age—a process made difficult as a result of various and complex nature of those modifications. Each particular person’s epigenome reacts uniquely to their atmosphere, resulting in refined variations in how we age, which is why a one-size-fits-all strategy to learning getting old has at all times fallen quick. With the arrival of machine studying, we now have the aptitude to investigate immense quantities of information to uncover patterns in these epigenetic modifications.
The primary iterations of epigenetic clocks have been launched in 2013 by two separate analysis groups: Hannum et al. and Horvath et al.4546 Hannum et al. centered on growing their clock utilizing blood samples, whereas Horvath’s strategy utilized samples from a number of tissues, permitting for broader functions throughout various kinds of organic supplies. The Horvath clock’s edge was in its versatility, it was capable of predict organic age whatever the supply of cells. The era strategy of the Horvath clock requires two inputs for every pattern: the DNA methylation information for round 20,000 methylation websites throughout the genome and the age of the affected person from which the pattern was derived. Then by means of a number of iterations of machine studying magic the algorithm sifts by means of the noise and identifies methylation web site patterns that appear to be correlated with age. The Horvath clock particularly makes use of 353 particular websites, some positively correlated, some negatively correlated with age at totally different weights. Solely by these methylation websites the horvath clock is ready to predict their chronological ages with outstanding accuracy.
One side wherein the Horvath clock falls quick is that it was educated solely on chronological age to interpret complicated DNA methylation information. Thus, it should assume that the chronological age and organic age of the sufferers from whom the coaching DNA methylation information was obtained are equal. After the invention of the Horvath clock, different DNA methylation clocks have been developed to deal with a major limitation. The Horvath clock, educated solely on chronological age, depends on the idea that the chronological and organic ages of the coaching samples align completely—an assumption that’s not at all times legitimate. To offer a extra correct measure of organic getting old, newer era clocks, such because the DNAm PhenoAge, have shifted their focus. Not like the Horvath clock, the DNAm PhenoAge shouldn’t be educated solely on chronological age however on phenotypic markers of getting old corresponding to, blood glucose, blood albumin, white blood cell depend and extra.47 There are different clocks that additional construct upon this and practice on parameters corresponding to remaining lifespan.48 This strategy helps these newer clocks extra precisely mirror a person’s organic age by incorporating broader well being and lifespan indicators, thereby bettering predictions of getting old outcomes and permitting interventions to be utilized extra successfully.

Determine 4: The three epigenetic clocks—Horvath’s Clock (blue line), DNAm PhenoAge (inexperienced line), and Hannum’s Clock (crimson line)—of their capacity to find out age acceleration and prediction capabilities throughout varied conditions. Horvath’s Clock excels in estimating age precisely throughout a number of tissues and age teams, together with supercentenarians however underperforms in phenotypic modifications like smoking and time to dying (left facet). DNAm PhenoAge is famous for its robust predictive accuracy for mortality, its correlation with smoking standing, and hyperlinks to markers of immune getting old. Hannum’s Clock, primarily used for blood samples, is efficient in predicting lifespan. Nonetheless, each carry out underneath the multi-tissue clock with regards to fundamental age from varied ages and tissues (proper facet). “AA” signifies age acceleration, with ‘AA blood’ particularly measuring it in blood samples. Sourced from Horvath et al.49
Moreover DNA methylation clocks, scientists have explored varied different epigenetic clocks corresponding to, primarily based on histone modifications and DNA accessibility.5051 Whereas these various approaches are intriguing and add depth to our understanding of getting old, they don’t but have the identical stage of widespread validation or intensive analysis backing that DNA methylation clocks do. Because it stands, DNA methylation clocks are probably the most trusted and well-established instruments within the discipline, celebrated for his or her constant accuracy in predicting organic age and evaluating the effectiveness of getting old interventions.
Whereas epigenetic clocks are revolutionary in getting old analysis, they face a number of limitations and challenges. One main concern is their reliance on the idea that chronological and organic ages align, which isn’t at all times true resulting from particular person life-style and well being variations (this downside is addressed with newer era clocks). In addition they are typically educated on particular pattern varieties, which can not characterize your complete inhabitants or all tissue varieties, limiting their common applicability. Moreover, the organic mechanisms underlying the epigenetic modifications these clocks measure usually are not totally understood, complicating their use in scientific settings the place the trigger and impact of getting old have to be clear.52
Conclusion
In conclusion, the intricate relationship between epigenetics and getting old is unveiling new layers of complexity in our understanding of organic getting old processes. Ongoing analysis on this space not solely deepens our grasp of how epigenetic mechanisms affect getting old but additionally brings forth progressive instruments like epigenetic clocks. These instruments maintain the potential to foretell and even perhaps modify the getting old trajectory. Within the subsequent article, we are going to discover present epigenetic interventions, these underneath growth, and potential future methods.